Technical Review No. 262, February 2023
Ben Cordingley, Amy Rinaldo, Caroline Bartel
Sections of this article were previously published in Aust. NZ. Grapegrower Winemaker (Cordingley 2022) and are reproduced here with permission from the publisher, Winetitles.
Brettanomyces bruxellensis (‘Brett’) is a yeast commonly found in wineries, which has the potential to cause significant spoilage in wines through the production of volatile phenol compounds. These compounds impart characteristics often described as ‘horsy, ‘Band-Aid’, ‘barnyard’ and ‘sweaty saddle’. Collectively these characters are often known as ‘Brett character’.
There are a range of strategies that winemakers can adopt to decrease the risk of spoilage from Brettanomyces. These usually work best when used in combination and include SO2 usage, pH management and rigorous winery hygiene. More detail about these mitigation and control strategies can be found in the AWRI fact sheet Controlling Brettanomyces during winemaking. In conjunction with control strategies, regular monitoring for Brettanomyces is recommended to allow early detection before volatile phenols have a negative sensory impact on wine. A range of options are available to test wines for the presence of Brettanomyces.
Testing methods
Most available testing methods for Brettanomyces fall into two main categories:
- Culture-dependent methods where Brettanomyces cells from a wine sample are cultured/grown on agar plates in the laboratory
- Culture-independent methods where Brettanomyces cells are detected by other means. This includes molecular tests using polymerase chain reaction (PCR) to detect Brettanomyces -specific DNA sequences, as well as flow cytometry methods that automate the identification and counting of Brett cells in a wine sample.
Each testing method works differently and can generate different results for the same sample due to the detection of different forms of Brettanomyces cells that can exist in a wine. A sample may contain a culturable population of viable Brettanomyces cells capable of growth. Several studies have reported that Brettanomyces can also exist in a dormant or viable but non-culturable (VBNC) state (Capozzi et al. 2016). In this theoretical state, live Brettanomyces cells may be metabolically active and capable of producing volatile phenols but temporarily unable to multiply (Nunes de lima et al. 2021). A sample may also contain dead Brettanomyces cells that contain detectable DNA, as well as free (extracellular) Brettanomyces DNA released during cellular lysis of dead cells.
Selective plating
The plating of wine samples onto nutrient agar is considered the gold standard in Brettanomyces detection. A volume of wine (commonly 50-100 mL) is filtered through a 0.45 µM membrane filter, which is then placed onto an agar that selectively allows Brettanomyces to grow while inhibiting most other yeast and bacteria. Wallerstein Nutrient agar (WL), supplemented with 100 mg/L cycloheximide, 50 mg/L chloramphenicol and 500 mg/L biphenyl is an example. As some Brettanomyces strains are slow growing, plates should be incubated for 10 days before results are reported. Any growth is analysed using a microscope to identify characteristics that indicate presumptive Brettanomyces. Selective plating is a highly sensitive method, with a limit of detection of 1 cell/volume of wine filtered. Only viable, culturable cells are detected using this method. VBNC and dead cells are not detected by culturing. Though this method is selective for Brettanomyces, it is not exclusive to Brettanomyces, and so false positives are possible if wild yeasts with similar morphology to Brettanomyces are present. Confirmation of Brettanomyces can be performed using a DNA-based method or sequencing.
PCR-based testing
Polymerase chain reaction (PCR) is a molecular test that uses enzymes to make copies of Brettanomyces-specific DNA. Brettanomyces DNA is recognised by synthetic molecules called primers that allow a part of Brettanomyces DNA to be amplified over multiple heating and cooling cycles. The number of Brettanomyces cells present in the sample is proportional to the amount of DNA that results from amplification. Conventional PCR methods have a predetermined number of amplification steps followed by a semi-quantitative estimation of the resulting amount of Brett DNA produced in the reaction. Quantitative PCR methods count the number of amplification steps needed to reach a threshold amount of Brett DNA that is detectable by the instrument. The number of amplification cycles required to reach this threshold level is used to precisely determine the number of Brett DNA copies in the sample prior to any amplification.
Conventional PCR technologies generally detect all intact Brettanomyces DNA from living and dead cells, and also free extracellular DNA. Some wines may contain high numbers of dead Brett cells that would still be included in a PCR-based cell count. The portion of dead cells present in a sample could depend on if there had been a treatment to kill Brett and how recently this treatment was applied. The time required for dead Brett cells to break down to the extent that their DNA is not detected by conventional PCR is unclear but may depend on wine conditions.
Affinity Labs offers the VeriflowTM PCR method to analyse for Brettanomyces cells in wine. This method has high specificity for Brettanomyces and does not cross react with other species that may be present in wine. The limit of detection is 10 cells/mL of wine. As a PCR-based method, it does not differentiate between live cells, dead cells and DNA present in the wine. Veriflow is rapid, and results are obtained within one day.
Some newer PCR methods do not detect dead cells and only detect viable and VBNC Brettanomyces. These methods exclude DNA from dead cells by making use of specialised dyes that are not able to enter viable cells but are able to enter dead cells. These dyes integrate within the DNA structure to prevent amplification by PCR (Navarro et al. 2020).
Flow cytometry
More recently, flow cytometry-based methods have become available that directly count the individual Brettanomyces cells in a sample. These work by creating a thin stream of the sample that flows past several detectors allowing the identification of Brettanomyces based on its physical properties or the binding of specific DNA probes. Flow cytometry techniques often use fluorescent dyes that selectively stain either living or dead Brettanomyces cells, so it is possible to distinguish between viable populations (potentially including VBNC Brett) and dead Brettanomyces cells.
Affinity Labs offers a flow cytometry-based rapid detection method, the Sysmex CyFlow™ BrettCount system. This method uses a mixture of fluorescent gene probes targeting different regions of the 16s ribosomal RNA of Brettanomyces cells to rapidly detect and quantify them in wine. This method has high specificity for Brettanomyces and does not cross react with other species that may be present in wine. The limit of detection is 100 cells/mL. Only viable cells are detected using this method, though some cells that are not culturable under laboratory conditions may also be detected. BrettCount is rapid, and results are obtained within one day. Table 1 provides a summary of the three methods offered by Affinity Labs.
Table 1. A comparison of the Brettanomyces bruxellensis detection methods offered by Affinity Labs
| Method | Volume required | Turnaround time (days) | Limit of detection | Detects dead cells | Specific to Brettanomyces bruxellensis | Quantitative | Suitable sample types |
| Selective plating | Up to 50 mL | 10 | 1 cell /50 mL | n | n | y | Wine, beer, other beverages |
| Flow cytometry | At least 1 mL | 1 | 100 cells /mL | n | y | semi | Wine, beer |
| PCR | At least 25 mL | 1 | 10 cells /mL | y | y | n | Wine |
Other methods
Rapid molecular tests using LAMP technology (loop-mediated isothermal amplification) have also been developed for the detection of Brettanomyces (Hayashi at al. 2007). This technology works similarly to PCR to amplify specific DNA sequences. The main difference is that LAMP uses a complex set of multiple primers that identify and amplify a Brettanomyces -specific DNA sequence at a constant temperature. LAMP is a rapid testing method that gives qualitative results (positive or negative) rather than a quantitative result expressed as the number of cells per mL of wine.
Method comparison
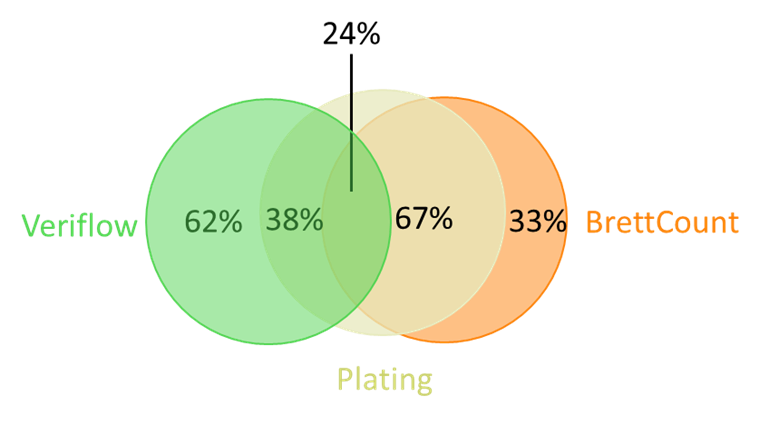
Affinity Labs recently conducted a comparative study on 21 Brettanomyces-positive wines to compare the three different detection methods it offers (selective plating, Veriflow and BrettCount). Wines were initially determined to by positive by Veriflow analysis and then analysed by both plating and BrettCount. Comparative analysis demonstrated that Veriflow results correlated with plating 38% of the time and BrettCount results correlated with plating 67% of the time (Figure 1). Differences in results between Veriflow and plating were attributed to the presence of residual DNA and/or dead cells in the wine sample. Differences in results between BrettCount and plating were attributed to cells being below detection limits and/or the presence of non-culturable cells. Overall, BrettCount was found to correlate most closely with selective plating, the current ‘gold standard’ for the detection of Brettanomyces cells in wine.
Figure 1. A summary of the consistency of results between the three methods. Twenty-one wines were determined to be positive for Brettanomyces by Veriflow. Wines were then analysed by BrettCount and selective plating. Veriflow results agreed with plating 38% of the time. BrettCount results agreed with plating 67% of the time. All three methods agreed 24% of the time.
Implications of a positive Brettanomyces detection
Brettanomyces cells can multiply from very low densities to result in a wine containing high levels of volatile phenols. Detection of any number of Brettanomyces cells should therefore prompt winemaking interventions to inactivate or remove the cells if a ‘Brett character’ is to be avoided. Wine conditions and the specific Brettanomyces strain can influence the production of volatile phenols, meaning that the cell density is not a direct indicator of the level of potential ‘Brett character’.
For further information about Brettanomyces bruxellensis detection or other technical winemaking or viticulture questions, contact the AWRI helpdesk on (08) 8313 6600 or helpdesk@awri.com.au.
For further information on testing for Brettanomyces bruxellensis in wine or other beverages, contact the Affinity Labs Applied Biosciences team on 08 8313 0444 or customerservice@affinitylabs.com.au.
References
Capozzi, V., Di Toro, M.R., Grieco, F., Michelotti, V., Salma, M., Lamontanara, A., Russo, P., Orrù, L., Alexandre, H., Spano, G. 2016. Viable But Not Culturable (VBNC) state of Brettanomyces bruxellensis in wine: New insights on molecular basis of VBNC behaviour using a transcriptomic approach. Food Microbiol. 59: 196-204.
Cordingley, B. 2022. Ask the AWRI: Techniques to detect Brettanomyces before it’s too late. Aust. N.Z. Grapegrower Winemaker (702): 70-71.
Hayashi, N., Arai, R., Tada, S., Taguchi, H., Ogawa, Y. 2007. Detection and identification of Brettanomyces/Dekkera sp. yeasts with a loop-mediated isothermal amplification method. Food Microbiol. 24(7-8): 778-785.
Navarro, Y., Torija, M.J., Mas, A., Beltran, G. 2020. Viability-PCR allows monitoring yeast population dynamics in mixed fermentations including viable but non-culturable yeasts. Foods. 9(10): 1373.
Nunes de Lima, A., Magalhães, R., Campos, F.M., Couto, J.A. 2021. Food Microbiol. 93: 103617.
Acknowledgement
The AWRI’s communications and the AWRI helpdesk are supported by Wine Australia, with levies from Australia’s grapegrowers and winemakers and matching funds from the Australian Government. The AWRI is a member of the Wine Innovation Cluster in Adelaide, SA.
Sysmex Australia is acknowledged for funding the comparative study of different Brettanomyces detection methods.

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.

