Bioprospecting Australian microbial genetic diversity
Project summary
An important aspect of terroir, particularly where spontaneous fermentations are performed, is very likely to be related to differences in wine microbiota. Focused microbiological research has shown that both vineyards and spontaneous fermentations contain diverse mixtures of microbial species (often with species being represented by multiple strains). However, the inability to transition this traditional methodology to efficiently and accurately assess the large numbers of samples required to tackle such a complex problem, has so far limited subsequent insights into this important question. This lack of information is therefore a major impediment to the exploitation of native microbial germplasm and spontaneous fermentation, by the Australian wine industry.
However, recent advances in metagenomics (genomic sequencing of mixed microbial communities), can address these issues by providing detailed identification of the species, and their proportions which comprise complex microbial mixtures, in a high throughput manner. In addition, by adopting and developing new genomic approaches, such as single cell high throughput sequencing, the genetic make-up of individual strains within these mixtures can also be obtained, to provide direct links between novel genetic and phenotypic characteristics. This type of metagenomic analysis has been initiated and refined for studies of wine fermentation at the AWRI, and now provides the technical platform to answer important questions regarding Australian wine microbial terroir.
This project builds on an existing project where the contributions of various species of fungi, yeast and bacteria are being elucidated in vineyard-to-wine time course experiments, using samples sourced from the spectrum of Australian wine regions, highlighting both the temporal and geographic dynamics of the microbial populations. Once these regional differences are identified, key microbes will be isolated in pure culture to assess their contributions to the unique terroir or wine style(s) of the region. In addition to regional wine isolates sourced from the metagenomics project, wild yeast strains will be sought from non-winemaking areas of Australia, as potential reservoirs of new and desirable winemaking characteristics, and uniquely Australian germplasm.
The thousands of strains that are isolated by this project, in addition to those already present in the AWRI Wine Microorganism Culture Collection, will be then assessed by high-throughput screening for desirable winemaking properties such as the production of key aroma compounds, the production of enzymes, and the ability to produce lower alcohol concentrations.
In addition, common winemaker interventions in wild fermentations will be explored by subjecting fermentations to various external factors (e.g. sulfite, oxygen, temperature, and/or high solids juice), to determine if it is possible to rationally influence or shape the performance or style of wild fermentations.
Strains that exhibit desirable phenotypes will progress to laboratory-scale fermentation and potentially to winery-scale and external trials, depending on their behaviour. These characterised strains will therefore provide a key resource for wineries to enhance the expression of regional terroir through inoculation of existing microbes either in the vineyard or within individual ferments.
Latest information
Mapping microbial communities in a winery
For the wine sector, metagenomics has provided detailed knowledge of the microbial diversity present on grapevines, in vineyard soil and in grape must. Food processing facilities, including wineries, are designed to not only ensure a consistent product, but also one that is safe for consumption. Despite careful hygiene, these facilities contain thriving microbial ecosystems which originate from raw substrates or other materials. In the case of the winery environment, grapes bring different bacterial and fungal populations, depending on variety, origin and harvesting practices. Although some resident microorganisms may not negatively affect the final product, spoilage microorganisms can be detrimental for quality, generating substantial economic losses.
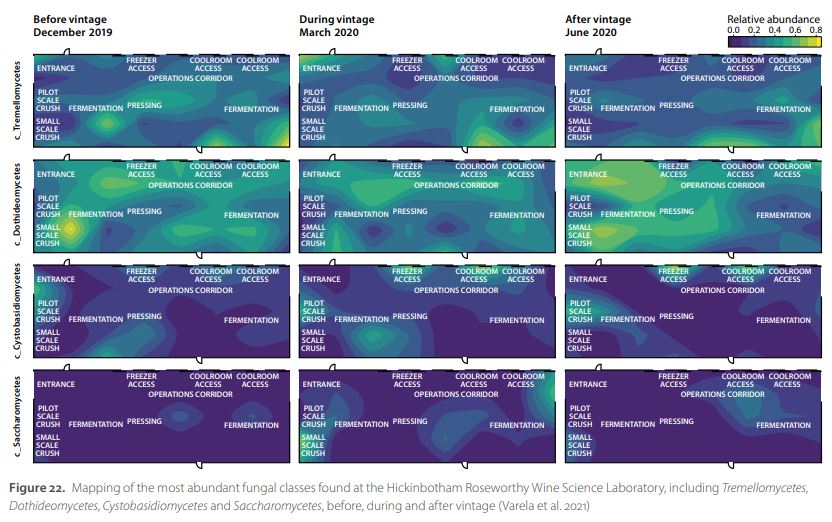
As a proof-of-concept, metagenomics techniques were used to map the microbial communities at the Hickinbotham Roseworthy Wine Science Laboratory on the Waite Campus, before, during and after the 2020 vintage (Varela et al. 2021). Resident bacterial and yeast populations changed over time, with both relative abundance and location within the winery varying according to sampling date. Vintage, the period when grapes are being processed in the winery, caused most of the changes in microbial structure, particularly for bacterial populations. Although fungal communities also changed during vintage, these changes were smaller than those observed for bacterial populations. The exception to this was Saccharomycetes, which showed approximately four times higher relative abundance during vintage than prior to this period. Mapping the spatial distributions of the microbial populations identified the main locations that harboured these resident microorganisms (Figure 22). This also included the wine spoilage genera Lactobacillus, Acetobacter, Gluconobacter and Brettanomyces, which were found mainly after vintage. Thus, the methodology described here can provide crucial information to wineries and other food processing facilities, helping to identify locations that need thorough disinfection and evaluate the efficacy of the existing sanitation practices.
Reference
Varela, C., Cuijvers, K., Borneman, A. 2021. Temporal comparison of microbial community structure in an Australian winery. Fermentation 7(3): 134.
Project Contact
Anthony Borneman, Cristian Varela

