Management and optimisation of the AWRI Wine Microorganism Culture Collection
Project summary
The AWRI Wine Microorganism Culture Collection (AWMCC) contains over 10,000 yeast and bacterial strains including yeast genome deletion libraries, proprietary strains, and natural isolates from Australian wineries. In addition, the collection is a largely untapped resource of microbial diversity which, if properly characterised, has the potential to greatly enhance microbial strain development projects.
Ongoing use of the culture collection is expected for future Wine Australia-funded projects, which will lead to the generation and/or isolation of many novel yeast and bacteria. Efficient use of the collection will require a reorganisation of storage facilities (including the off-site duplicate collection), and an update of the existing collection database into a format that is amenable to high throughput screening and characterisation. In addition, quality control (QC) of new depositions and existing strains within the collection, will be required on an ongoing basis, and technological advances may provide opportunities to improve existing QC regimes. Once the collection has been organised for high throughput screening, large scale genotyping and phenotyping will be feasible.
Latest information
Identification, storage and distribution of microbial strains
In 2020/2021, a total of 224 individual yeast and bacterial strains were submitted to the AWMCC by researchers and wineries. During the year, the AWMCC distributed 513 microbial strains from cryogenic stocks to AWRI researchers, wineries and external research partners.
Microorganism species identification is a dynamic area, and species relationships and identities are constantly being updated, so the collection is periodically rationalised with new species designations. The goal is to identify the isolates correctly and efficiently to make the process of screening yeast or bacteria for researchers more relevant, and to be able to provide appropriate cultures for winemakers. Historically, techniques such as microscopic analysis of cell morphology, biochemical and physiological testing have been used to identify microorganisms. However, such morphological and biochemical techniques are increasingly being demonstrated to be prone to inaccuracies. The current method of molecular characterisation is undergoing development to take into account next generation sequencing technologies, and the ability to identify large numbers of organisms via a plate-based system rather than a one-at-a-time methodology. This will have the potential to increase the speed and cost efficiency of molecular identification and will create a curated database of cultures that can be used for a range of purposes.
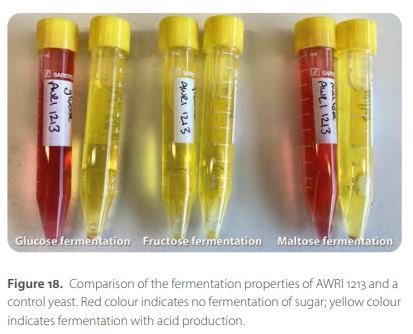
During the year, a number of strains with unusual fermentation properties were characterised that have potential for use in wine production and other industries. These included a hybrid Saccharomyces yeast with high sorbitol fermentation potential, which may be useful in the pharmaceutical industry, and a yeast isolate (AWRI 1213) that appears to ferment fructose but not glucose, which may be useful in low-alcohol beverage production (Figure 18).